Digital Connectivity Architecture of the Brain : Connectomics and its Application in Neurosurgery
- BrainSightAI Team
- Oct 25, 2022
- 4 min read
Author: Janova anbarasi
The twentieth century clinical neuroscience research in the last decade has centred around identifying, classifying and localizing individual regions of the brain to uncover how complex regional specializations give rise to complex behaviour and cognition. Certain specialized regions in the brain become integrated and evolves with development and this constitutes complex brain connectivity. Human connectomics is a new area of study made possible by recent developments in non-invasive neuroimaging that allow for the measuring of connections across dispersed brain regions while they are still alive.
Connectomics has become crucial to study because neural insults occurring in one brain region does not necessarily emerge from damage to discreate and specialized brain regions but involves other regions of the brain too. This demonstrates a balanced, complex, and precisely coordinated dynamics across multiple spatio-temporal scales. A network displays connectedness, which harbours the characteristics that a dysfunction can quickly spread across connected elements, setting off a pathological cascade that can affect substantial portions of the system. Axonal and synaptic connections in the brain can serve as vehicles for the spread of neurological disease processes. An example of this would be the speed with which focal epileptogenic activity transforms into generalized seizures, the spatially distributed activation changes brought on by regionally localized ischaemic insults and the slow progression of pathology in degenerative diseases with focal onset like Huntington disease, Parkinson disease, and other types of neurodegeneration disorders [1,2].
The fundamental objective of connectomics is the complete comprehensive analysis of brain connectivity, including mapping throughout all dimensions, from the macro-scale of the brain to the micro-scale of the individual synaptic connections between neurons and interregional pathways. Two methods are frequently used to map interregional connections in living organisms which are diffusion tractography and resting state functional MRI (Magnetic Resonance Imaging). They both employ MRI, but their approaches are significantly different. The core aim of diffusion tractography is to reconstruct, millimetre by millimetre, the paths taken by axon bundles as they move through the white matter of the brain. Until recently, only post-mortem dissection or invasive tracing in non-human animals could be used to study such complicated white matter anatomy. The anisotropic diffusion of water in and around axons is crucial to all diffusion tractography research. Although molecules that are freely diffusing will diffuse in all directions equally, the existence of semi-permeable tissue barriers may impede diffusion in some orientations but not others Axonal membranes and myelin sheaths of the brain’s white matter prevent diffusion perpendicular to the axon, leaving diffusion to occur most quickly along the axon. This mechanism of orientational dependence of diffusion is referred to as anisotropy. This anisotropy is measurable and the peak diffusion orientations in each image voxel using diffusion weighted MRI. Nevertheless, these voxels cover 1.5 X 1.5 x 1.5 mm3, tissue cubes that may each contain tens of thousands of axons. The orientations of bundles of coherently directed axons travelling through the voxel are therefore related to the diffusion orientations that are measured. Diffusion tractography reconstructs these bundles and subsequently interregional connections by following their orientations voxel-to-voxel through the white matter.
In contrast, fMRI does not directly evaluate brain connections, they instead represent connection as statistical dependencies between patterns of activity in the grey matter. At rest, regional brain activity fluctuates at low frequencies, that cross-correlate far flung brain areas. [3,4]. Certain network nodes are identified and then functional connection (network edges) between these nodes are estimated. During brain parcellation, nearby voxels (3D pixels) are grouped based on how similar their timeseries are in order to produce nodes. This is known are a “hard parcellation” because it often produces a high number of non-overlapping parcels with a single contiguous collection of voxels in each parcel or node. Resting state fMRI is known to be very informative about brain organisation, the variability of functional connections between individuals is predictive of genetic variability. [5,6].
In order to perform safe and effective neurosurgery, the knowledge of functional anatomy is essential. Connectome analysis provides intuitive modelling of lesions and plasticity which is a practical option for neurosurgeons. Connectome analysis was applied to patients with brain tumours to characterise overall network topology and individual patterns of connectivity alterations. Brain parcellation was done by parcellating the brain into anatomical regions, which initiated complex network analysis. The most significant correlations between respective wavelet decomposed time-series were retained and hence connections identified.
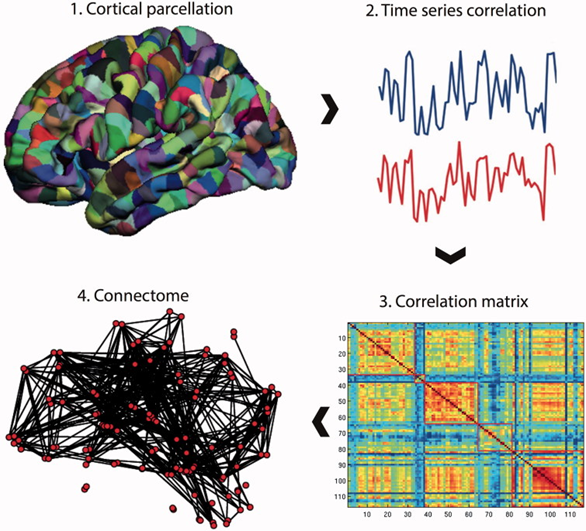
Fig.1 Building a connectome
Source: Micheal G. et al., 2016
A connectome analysis is performed using resting-state fMRI data as an example, but similar procedures can be used with data from DTI or EEG/MEG. The brain is initially divided into many areas (referred to as parcels) that make up the network nodes using a template. These nodes are what make up a matrix's rows and columns. Each node's edges are represented by entries in the matrix, which are created by recording a statistic (such the Pearson correlation co-efficient) between each node's resting-state fMRI time series [7].

Fig.2 Digital Brain Map
Source: https://www.o8t.com/about
This comprehensive map of the brain also referred to as the ‘wiring diagram” has been constructed completely and successfully for roundworm C. elegans. Research has also managed to construct partial connectomes of retain of mouse and its primary visual cortex. The mapping of the human brain is connectomics' ultimate goal and to make neurosurgery more feasible, practical and innovative. It gives advantage for pre-surgical planning by identifying critical regions at risk and networks that should be preserved. Future modelling of functional plasticity and recovery from cognitive disorders could be done using such digitally available connectomics data [8].
References:
Fornito, A., Zalesky, A., & Breakspear, M. (2015). The connectomics of brain disorders. Nature Reviews Neuroscience, 16(3), 159-172.
Hagmann, P., Grant, P. E., & Fair, D. A. (2012). MR connectomics: a conceptual framework for studying the developing brain. Frontiers in systems neuroscience, 6, 43.
Behrens, T. E., & Sporns, O. (2012). Human connectomics. Current opinion in neurobiology, 22(1), 144-153.
Johansen-Berg H, Behrens TE. Just pretty pictures? What diffusion tractography can add in clinical neuroscience. Curr Opin Neurol. 2006;19(4):379-385. doi:10.1097/01.wco.0000236618.82086.01.
Behrens, T. E., & Sporns, O. (2012). Human connectomics. Current opinion in neurobiology, 22(1), 144–153. https://doi.org/10.1016/j.conb.2011.08.005.
Smith SM, Vidaurre D, Beckmann CF, et al. Functional connectomics from resting-state fMRI. Trends Cogn Sci. 2013;17(12):666-682. doi:10.1016/j.tics.2013.09.016
Hart MG, Price SJ, Suckling J. Connectome analysis for pre-operative brain mapping in neurosurgery. Br J Neurosurg. 2016;30(5):506-517. doi:10.1080/02688697.2016.1208809
https://operativeneurosurgery.com/doku.php?id=connectome&rev=1564044283




Comments